ARCANE

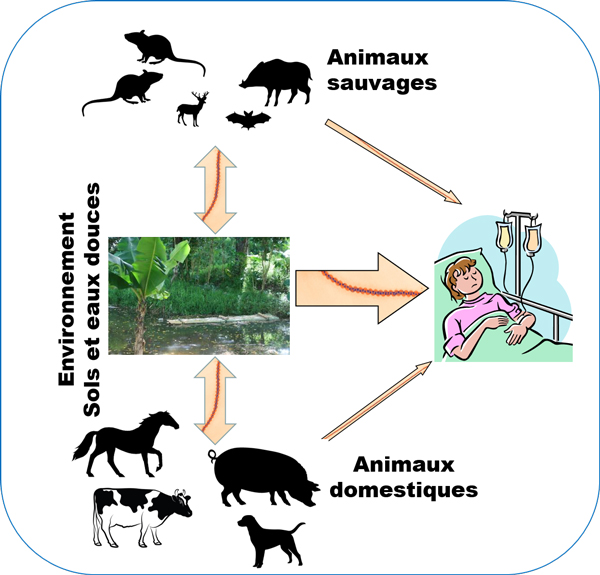
Rapid emergence and spread of pathogenic bacteria (multi)resistant to antimicrobials is a priority health issue. Despite its insularity, New Caledonia has not been spared by this pandemic and antibiotic resistance represents a major public health issue in the territory. The surveillance carried out until now is focused on clinical approaches and there is no study to date on the role of the environment in the selection and dissemination of antibiotic-resistant bacteria.
In this context, the ARCANE project proposes to study the resistance of bacterial populations in wastewater (from hospitals or the general population) and the receiving environment (coastal seawater).
From a public health point of view, this project will allow to draw up a representation of antibiotic resistance in the New Caledonian environment and to identify critical points amenable to control actions aiming at limiting the spread of antimicrobial resistance.
Alexandre Bourles
Pôle Bactériologie – Unité de Bactériologie Expérimentale

The rapid emergence and spread of pathogenic bacteria (multi)resistant to antimicrobials is a priority health issue. Despite its insularity, New Caledonia has not been spared from this pandemic: the territory was the host country of the first French isolate carrying the plasmid gene mcr-1 (conferring resistance to colistin). Other resistances are of increasing concern because they cause significant public health problems in the territory. For example, methicillin-resistant Staphylococcus aureus is spreading in the community with highly virulent strains and Carbapenem Resistant Enterobacteriaceae (CRE) are involved in epidemics in the Territorial Hospital.
The selection and transmission of these resistances are frequently investigated through clinical studies and it is only recently that the role of the environment as a source and pathway for the diffusion of antibiotic resistance has been recognized. Indeed, effluents from hospitals, industry, livestock or veterinary structures, wastewater treatment plants and urban/agricultural runoff are all potential sources of new contaminants (antibiotics, biocides, metals, resistance genes, resistant bacteria) that can be discharged into receiving environments. In addition to anthropogenic inputs, the high concentrations of metals found naturally can also add factors favoring the emergence and/or selection of antibiotic-resistant bacteria. Indeed, the genes for resistance to metals and antibiotics may be located on the same mobile genetic elements (co-resistance), or an identical mechanism may be involved in resistance to antibiotics and metals (cross-resistance).
Among the techniques aimed at highlighting these sources of contaminants and the risks to human health, epidemiology based on wastewater analysis via metagenomic approaches has been developed in recent years. These innovative approaches make it possible to identify bacterial species but also to quantify and describe thousands of genes (e.g. resistance or virulence) in a given sample.
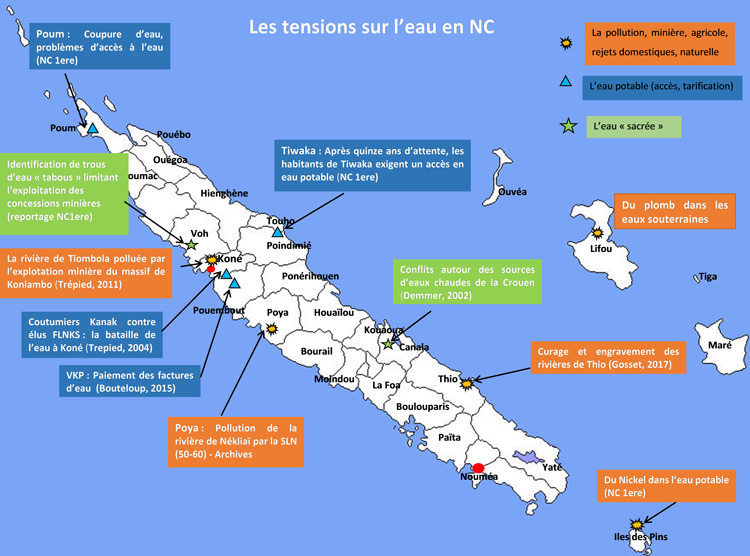
In New Caledonia, no study aimed at identifying resistant bacteria in wastewater, runoff or the receiving environment has been conducted to date. However, there is a real need in the territory since, during heavy rains for example, it is not uncommon to see bacterial contamination of bathing areas. Sanitary monitoring of bathing waters is carried out annually during the summer period (from December to April) by the New Caledonian Government's Department of Health and Social Affairs throughout the territory. In some municipalities , additional analyses are also carried out. This monitoring focuses on the enumeration of Escherichia coli and intestinal enterococci without assessing antibiotic resistance.
In this context, the objectives of the ARCANE project are to evaluate the dissemination of resistance genes and antibiotic-resistant bacteria in the environment, with an innovative integrated environmental approach. Thanks to the development of an innovative technology for monitoring metal and antibiotic concentrations, this project will also identify possible factors of anthropic or environmental origin likely to promote the selection, development and dissemination of resistant bacterial strains.
This project includes three CRESICA partners: Institut Pasteur of New Caledonia, New Caledonia Central Hospital CHT, and French National Research Institute for Sustainable Development IRD.The metagenomic part will be carried out in partnership with the Institut Pasteur in Paris and the assays of metals and antibiotics will involve the private company AEL. This project also involves the New Caledonian authorities and institutions responsible for health (DASS, DAVAR, Noumea municipality, Dumbéa municipality, SIVOM-VKP), as well as the Environment (South Province and North Province) for the different sampling sites, the health care institutions concerned by the study and the managers of the wastewater treatment systems selected.

-
Sampling
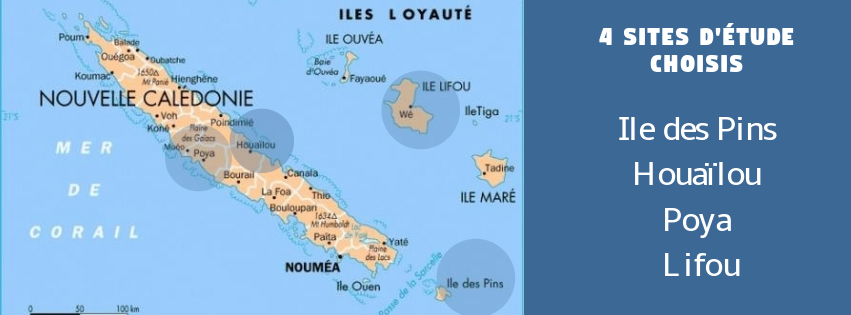
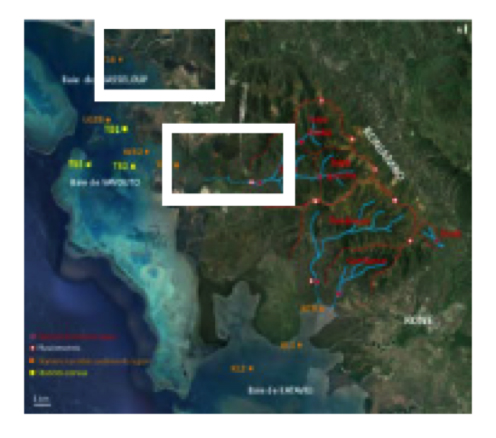
The monitoring will be carried out longitudinally during a 14-month sampling campaign (sampling every other month). The targeted sampling sites follow the path of wastewater: from the effluents of the healthcare structures to the receiving environment. Punctual water sampling as well as 24-hour sampling will be carried out on certain sites.
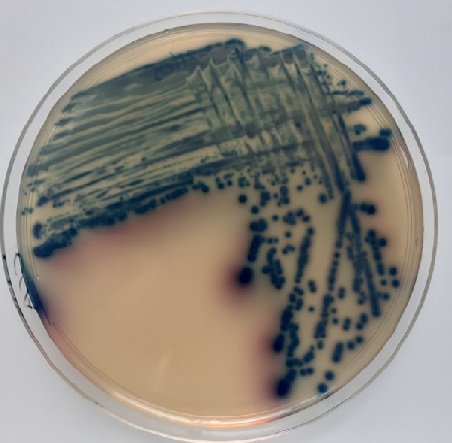
2. Culture
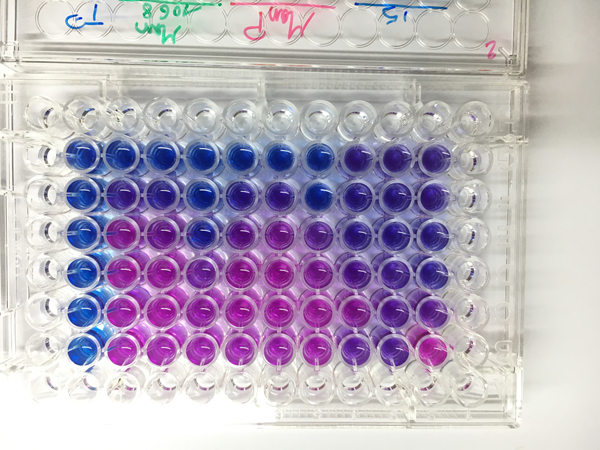
Concerning the isolations, a selection of environmental isolates, with a particular focus on Enterobacteriaceae producing (i) carbapenemases, (ii) extended spectrum beta-lactamases and (iii) high level of cephalosporinsase is carried out. The technique is based on direct plating on culture medium or filtration of water samples and deposition of membranes on selective agars to isolate β-lactam resistant Enterobacteriaceae. An identification by MALDI-ToF is performed and synergy tests allow to identify the resistance mechanism involved. These isolations will provide an overview of the resistance mechanisms found in cultivable bacteria in the environment. Some isolates will be subjected to whole genome sequencing to identify the repertoire of genes and mobile genetic elements involved in resistance to serve as a reference repertoire for metagenomics analyses. In parallel, complete genomes of clinical and hospital isolates of interest will be added to the database.
3. Metagenomic
For metagenomics, the proposed approach consists in collecting organisms and debris (from organisms) present in water by filtration at 0.22 µm. The DNA will be extracted and subjected to shotgun sequencing. This approach allows to gain a global view of the microbiome as well as its functional potential by identifying the genes and mobile genetic elements represented. The consensus sequences obtained after assembly of the sequencing reads will be aligned with referenced genomes for the identification of the species forming the microbiome. Comparisons of bacterial communities sampled in different environments as well as resistance profiles and their variations in time and space will be performed. The longitudinal approach from the source to the coastal waters will allow a detailed description of the circulation of resistant bacteria and resistance genes and to inform the possible sanitary risks.
4. Integrative monitoring of metals and antibiotics
An integrative technique for the analysis of trace metals and trace and ultra-trace antibiotics on thin film resin (Diffusive Gradient in Thin film; DGT) will be used in addition to classical assays on waters.
Les 28 et 29 mars 2022, le CRESICA organisait le séminaire "Au fil de l'eau" qui a permis de présenter l'état d'avancement du projet.
Visionnez la présentation fait lors du séminaire grâce à la vidéo du projet du projet ARCANE par A. Bourles.
Le 29 août 2023, le CRESICA organisait le séminaire de restitution "Au fil de l'eau" qui a permis de présenter les résultats du projet.
Brochure : "Des projets de recherche sur la thématique de l'eau en Nouvelle-Calédonie."
Visionnez la présentation faite lors du séminaire grâce à la vidéo du projet ARCANE par A.Bourles
Rapport final : projet ARCANE





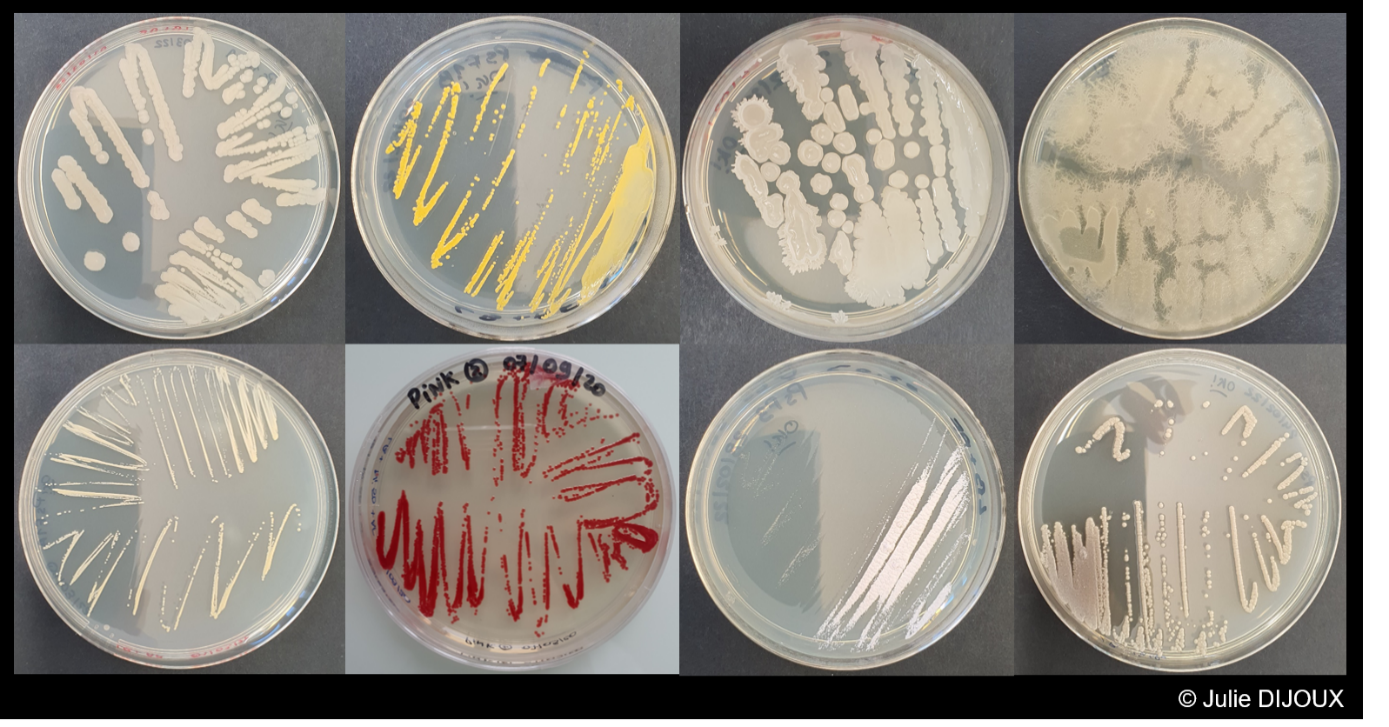
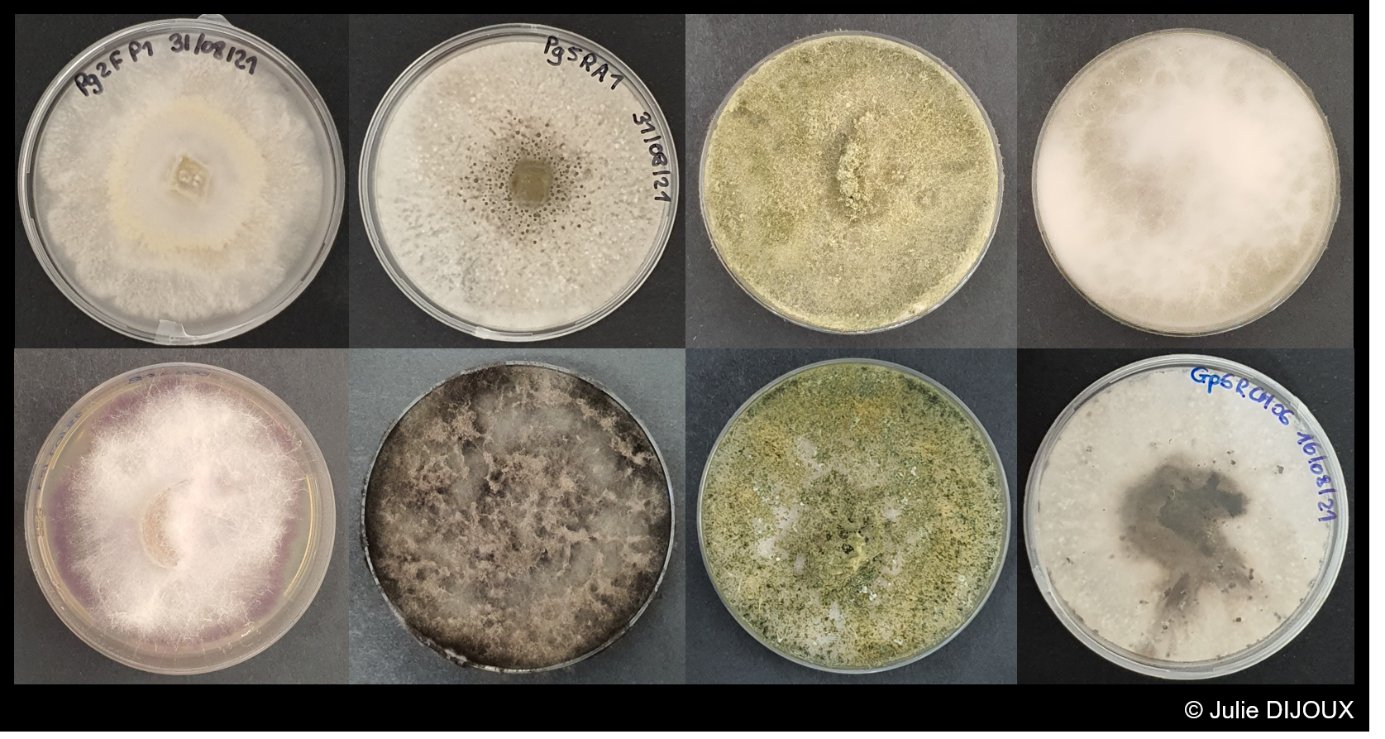





















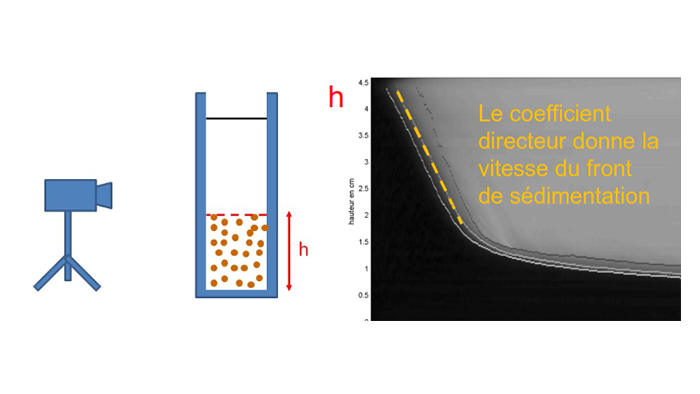



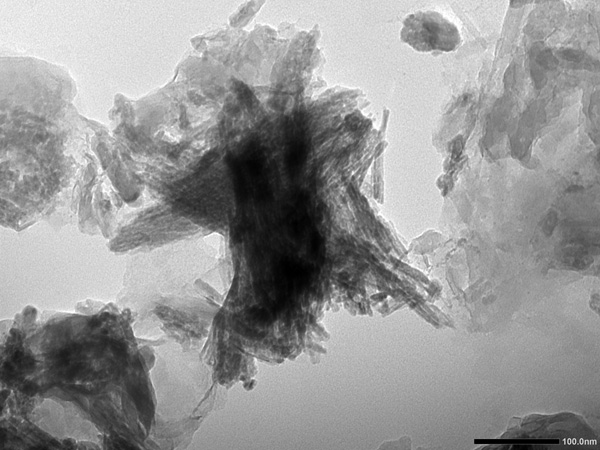

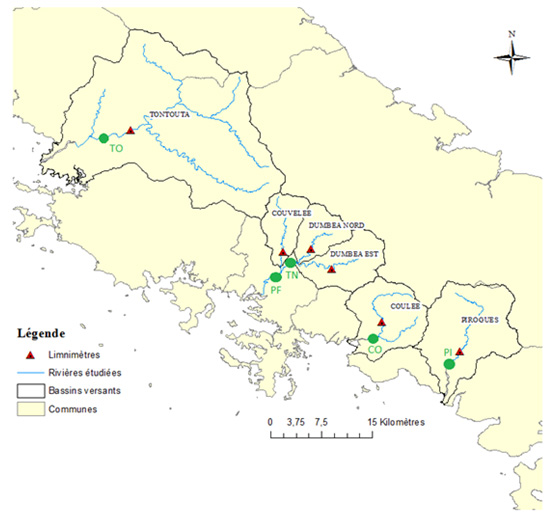
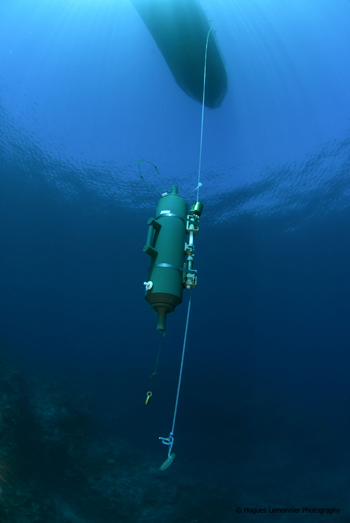




 The project is therefore structured around 5 major work packages spread over 3 years, with specific contributors assigned to each package.
The project is therefore structured around 5 major work packages spread over 3 years, with specific contributors assigned to each package.